Your job calculation is finished.
Main results
Metabolite information
Enzyme information
Kinetic parameters
| Enzyme | MVFDLKRIVRPK ... YFKETLYKLANE
MVFDLKRIVRPKIYNLEPYRCARDDFTEGILLDANENAHGPTPVELSKTNLHRYPDPHQLEFKTAMTKYRNKTSSYANDPEVKPLTADNLCLGVGSDESIDAIIRACCVPGKEKILVLPPTYSMYSVCANINDIEVVQCPLTVSDGSFQMDTEAVLTILKNDSLIKLMFVTSPGNPTGAKIKTSLIEKVLQNWDNGLVVVDEAYVDFCGGSTAPLVTKYPNLVTLQTLSKSFGLAGIRLGMTYATAELARILNAMKAPYNISSLASEYALKAVQDSNLKKMEATSKIINEEKMRLLKELTALDYVDDQYVGGLDANFLLIRINGGDNVLAKKLYYQLATQSGVVVRFRGNELGCSGCLRITVGTHEENTHLIKYFKETLYKLANE
|
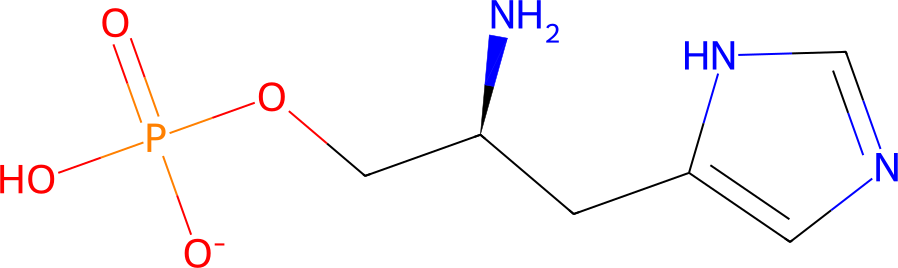
| Metabolite | InChI=1S/C6H ... -1/t5-/m0/s1
InChI=1S/C6H12N3O4P/c7-5(3-13-14(10,11)12)1-6-2-8-4-9-6/h2,4-5H,1,3,7H2,(H,8,9)(H2,10,11,12)/p-1/t5-/m0/s1
|
| How often was metabolite in training set? | 8 |
| Prediction score | 0.96 |
Based on experimental data in our database, the following proteins function as enzymes for the metabolite you entered:
| UniProt ID | Gene ID | Sequence | Copy |
|---|---|---|---|
| P07172 | HIS5 |
MVFDLKRIVRPK ... YFKETLYKLANE
MVFDLKRIVRPKIYNLEPYRCARDDFTEGILLDANENAHGPTPVELSKTNLHRYPDPHQLEFKTAMTKYRNKTSSYANDPEVKPLTADNLCLGVGSDESIDAIIRACCVPGKEKILVLPPTYSMYSVCANINDIEVVQCPLTVSDGSFQMDTEAVLTILKNDSLIKLMFVTSPGNPTGAKIKTSLIEKVLQNWDNGLVVVDEAYVDFCGGSTAPLVTKYPNLVTLQTLSKSFGLAGIRLGMTYATAELARILNAMKAPYNISSLASEYALKAVQDSNLKKMEATSKIINEEKMRLLKELTALDYVDDQYVGGLDANFLLIRINGGDNVLAKKLYYQLATQSGVVVRFRGNELGCSGCLRITVGTHEENTHLIKYFKETLYKLANE
|
|
| Q8NS80 | hisN |
MSKYADDLALAL ... GILHDETLDRLK
MSKYADDLALALELAELADSITLDRFEASDLEVSSKPDMTPVSDADLATEEALREKIATARPADSILGEEFGGDVEFSGRQWIIDPIDGTKNYVRGVPVWATLIALLDNGKPVAGVISAPALARRWWASEGAGAWRTFNGSSPRKLSVSQVSKLDDASLSFSSLSGWAERDLRDQFVSLTDTTWRLRGYGDFFSYCLVAEGAVDIAAEPEVSLWDLAPLSILVTEAGGKFTSLAGVDGPHGGDAVATNGILHDETLDRLK
|
|
| P06987 | hisB |
MSQKYLFIDRDG ... EGDTLPSSKGVL
MSQKYLFIDRDGTLISEPPSDFQVDRFDKLAFEPGVIPELLKLQKAGYKLVMITNQDGLGTQSFPQADFDGPHNLMMQIFTSQGVQFDEVLICPHLPADECDCRKPKVKLVERYLAEQAMDRANSYVIGDRATDIQLAENMGITGLRYDRETLNWPMIGEQLTRRDRYAHVVRNTKETQIDVQVWLDREGGSKINTGVGFFDHMLDQIATHGGFRMEINVKGDLYIDDHHTVEDTGLALGEALKIALGDKRGICRFGFVLPMDECLARCALDISGRPHLEYKAEFTYQRVGDLSTEMIEHFFRSLSYTMGVTLHLKTKGKNDHHRVESLFKAFGRTLRQAIRVEGDTLPSSKGVL
|
|
| P06986 | hisC |
MSTVTITDLARE ... QRVIDALRAEQV
MSTVTITDLARENVRNLTPYQSARRLGGNGDVWLNANEYPTAVEFQLTQQTLNRYPECQPKAVIENYAQYAGVKPEQVLVSRGADEGIELLIRAFCEPGKDAILYCPPTYGMYSVSAETIGVECRTVPTLDNWQLDLQGISDKLDGVKVVYVCSPNNPTGQLINPQDFRTLLELTRGKAIVVADEAYIEFCPQASLAGWLAEYPHLAILRTLSKAFALAGLRCGFTLANEEVINLLMKVIAPYPLSTPVADIAAQALSPQGIVAMRERVAQIIAEREYLIAALKEIPCVEQVFDSETNYILARFKASSAVFKSLWDQGIILRDQNKQPSLSGCLRITVGTREESQRVIDALRAEQV
|
|
| P0DI07 | HPA1 |
MGVINVQGSPSF ... TDVLMECLKQFY
MGVINVQGSPSFSIHSSESNLRKSRALKKPFCSIRNRVYCAQSSSAAVDESKNITMGDSFIRPHLRQLAAYQPILPFEVLSAQLGRKPEDIVKLDANENPYGPPPEVFEALGNMKFPYVYPDPQSRRLRDALAQDSGLESEYILVGCGADELIDLIMRCVLDPGEKIIDCPPTFSMYVFDAAVNGAGVIKVPRNPDFSLNVDRIAEVVELEKPKCIFLTSPNNPDGSIISEDDLLKILEMPILVVLDEAYIEFSGVESRMKWVKKYENLIVLRTFSKRAGLAGLRVGYGAFPLSIIEYLWRAKQPYNVSVAGEVAALAALSNGKYLEDVRDALVRERERLFGLLKEVPFLNPYPSYSNFILCEVTSGMDAKKLKEDLAKMGVMVRHYNSQELKGYVRVSAGKPEHTDVLMECLKQFY
|
|
| P38635 | HIS2 |
MHSHHSHSGDYS ... SQFVNDPFWANI
MHSHHSHSGDYSAHGTDPLDSVVDQVVNLNFHTYCLTEHIPRIEAKFIYPEEQSLGKNPEEVITKLETSFKNFMSHAQEIKTRYADRPDVRTKFIIGMEIESCDMAHIEYAKRLMKENNDILKFCVGSVHHVNGIPIDFDQQQWYNSLHSFNDNLKHFLLSYFQSQYEMLINIKPLVVGHFDLYKLFLPNDMLVNQKSGNCNEETGVPVASLDVISEWPEIYDAVVRNLQFIDSYGGAIEINTSALRKRLEEPYPSKTLCNLVKKHCGSRFVLSDDAHGVAQVGVCYDKVKKYIVDVLQLEYICYLEESQSPENLLTVKRLPISQFVNDPFWANI
|
|
| Q9K4B1 | hisN |
MPDYLDDLRLAH ... HDELLGYLNQRY
MPDYLDDLRLAHVLADAADAATMDRFKALDLKVETKPDMTPVSEADKAAEELIRGHLSRARPRDSVHGEEFGVAGTGPRRWVIDPIDGTKNYVRGVPVWATLIALMEAKEGGYQPVVGLVSAPALGRRWWAVEDHGAFTGRSLTSAHRLHVSQVSTLSDASFAYSSLSGWEEQGRLDGFLDLTREVWRTRAYGDFWPYMMVAEGSVDLCAEPELSLWDMAANAIIVTEAGGTFTGLDGRPGPHSGNAAASNGRLHDELLGYLNQRY
|
|
| Q6NPM8 | IMPL2 |
MLAQSHFFSKSF ... IHQQALESLEWH
MLAQSHFFSKSFDLIPPQSPALRSANPSLRISSSYSNSRLSFLSSSAIAVPVSRRRFCLTMASNSKRPNISNESPSELSDTELDRFAAVGNALADASGEVIRKYFRKKFDIVDKDDMSPVTIADQMAEEAMVSIIFQNLPSHAIYGEEKGWRCKEESADYVWVLDPIDGTKSFITGKPVFGTLIALLYKGKPILGLIDQPILKERWIGMNGRRTKLNGEDISTRSCPKLSQAYLYTTSPHLFSEEAEKAYSRVRDKVKVPLYGCDCYAYALLASGFVDLVIESGLKPYDFLALVPVIEGAGGTITDWTGKRFLWEASSSAVATSFNVVAAGDSDIHQQALESLEWH
|
Based on experimental data in our database, the following molecules are substrates for the enzyme you entered:
| Molecule ID | InChI String | Copy |
|---|---|---|
| CHEBI:57980 |
InChI=1S/C6H12N3O4P/c7-5( ... 2,10,11,12)/p-1/t5-/m0/s1
InChI=1S/C6H12N3O4P/c7-5(3-13-14(10,11)12)1-6-2-8-4-9-6/h2,4-5H,1,3,7H2,(H,8,9)(H2,10,11,12)/p-1/t5-/m0/s1
|
|
| CHEBI:16810 |
InChI=1S/C5H6O5/c6-3(5(9) ... 1-2H2,(H,7,8)(H,9,10)/p-2
InChI=1S/C5H6O5/c6-3(5(9)10)1-2-4(7)8/h1-2H2,(H,7,8)(H,9,10)/p-2
|
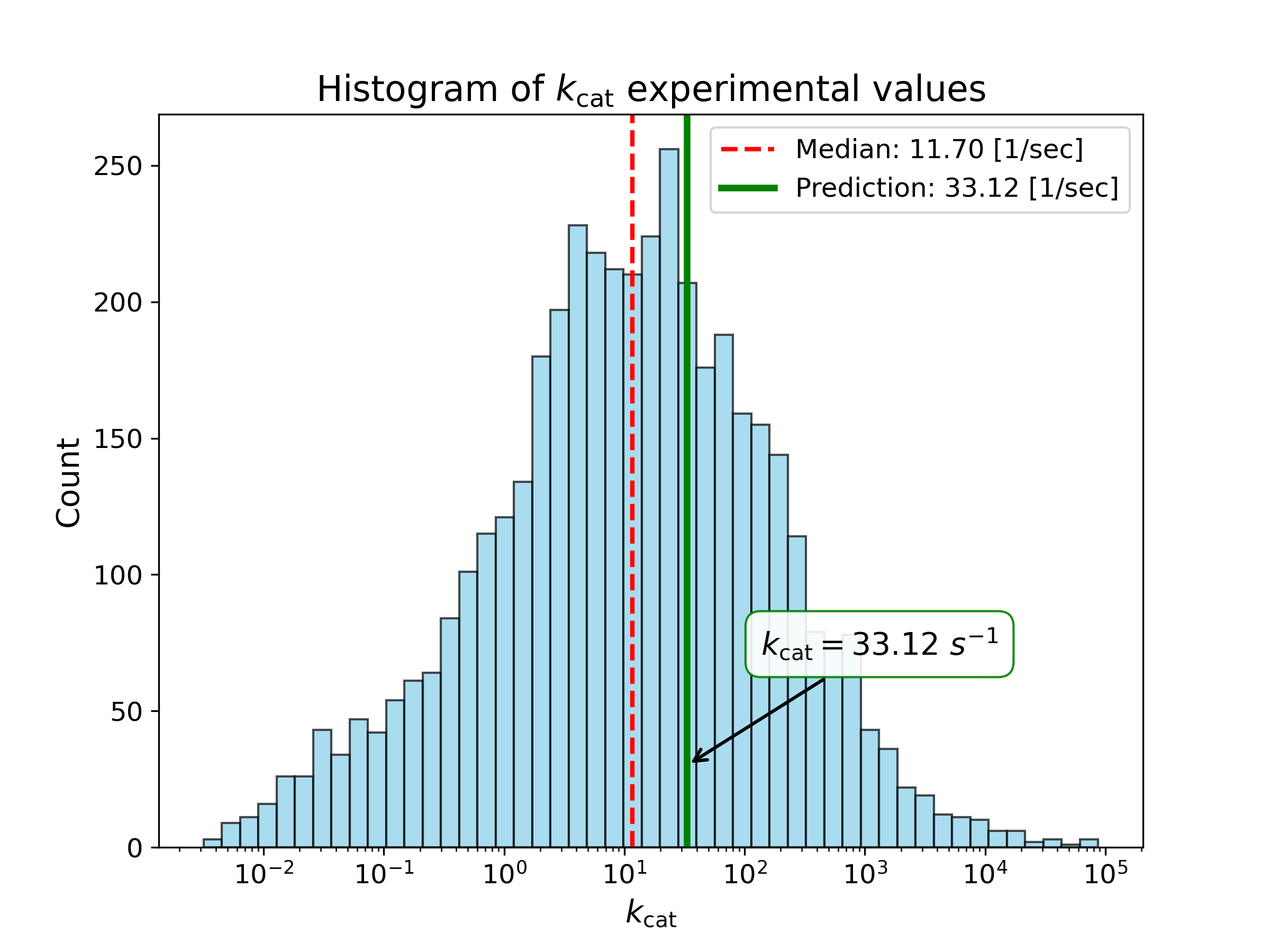
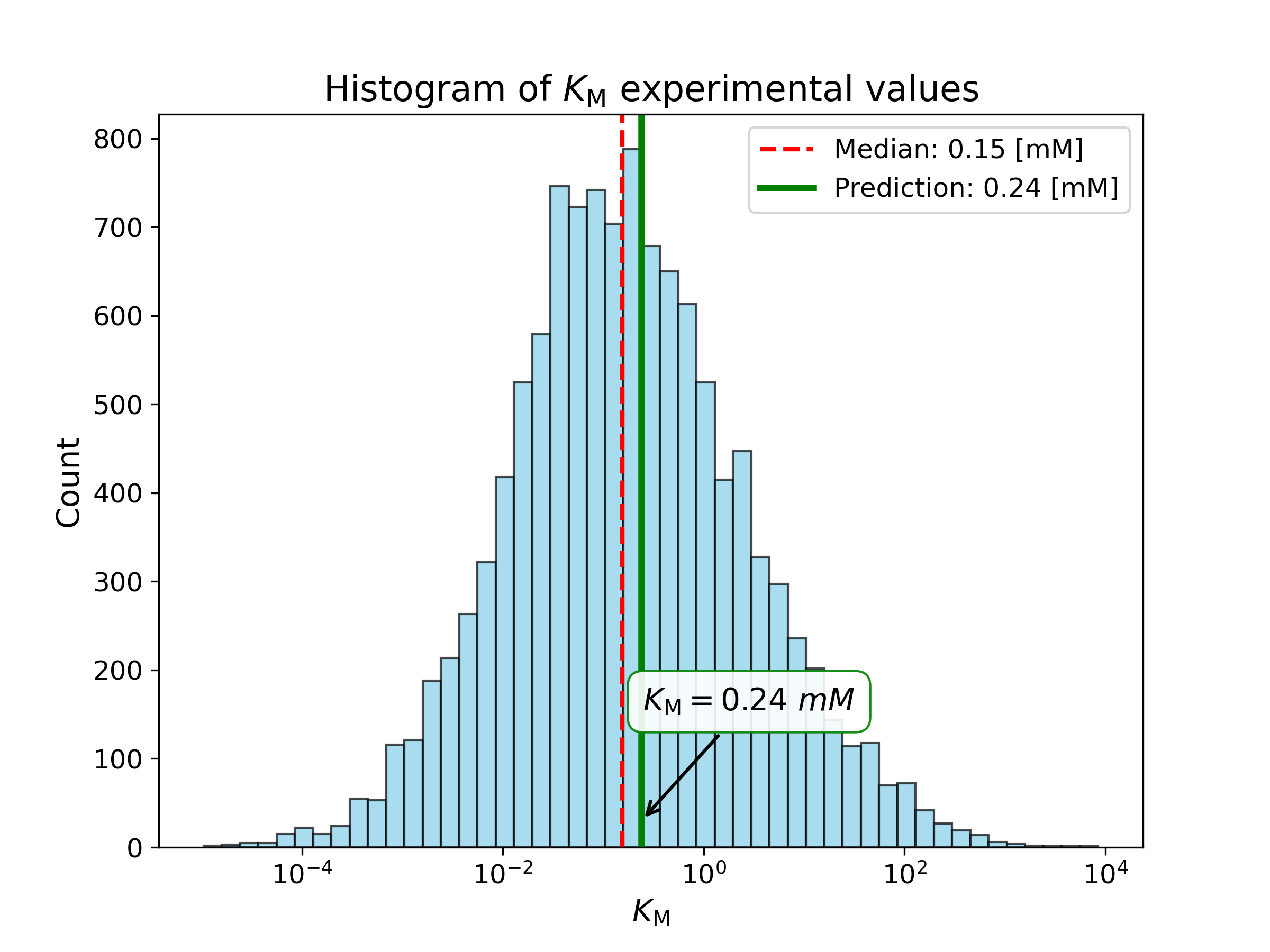
We computed the following kinetic parameters with our prediction model for the enzyme and enzyme-metabolite pair you provided:
| Parameter | Value | Model Input |
|---|---|---|
| Turnover Number |
33.12 [1/sec] | Enzyme |
| Michaelis constant KM | 0.24 [mM] | Enzyme-Metabolite-Pair |
 KM predictions are only meaningful if the ESP prediction score (see Main results) is above 0.5.
KM predictions are only meaningful if the ESP prediction score (see Main results) is above 0.5.